Platinum »
PDB 1uas-3co3 »
2k0u »
Platinum in PDB 2k0u: High Resolution Solution uc(Nmr) Structures of Oxaliplatin-Dna Adduct
Platinum Binding Sites:
The binding sites of Platinum atom in the High Resolution Solution uc(Nmr) Structures of Oxaliplatin-Dna Adduct
(pdb code 2k0u). This binding sites where shown within
5.0 Angstroms radius around Platinum atom.
In total only one binding site of Platinum was determined in the High Resolution Solution uc(Nmr) Structures of Oxaliplatin-Dna Adduct, PDB code: 2k0u:
In total only one binding site of Platinum was determined in the High Resolution Solution uc(Nmr) Structures of Oxaliplatin-Dna Adduct, PDB code: 2k0u:
Platinum binding site 1 out of 1 in 2k0u
Go back to
Platinum binding site 1 out
of 1 in the High Resolution Solution uc(Nmr) Structures of Oxaliplatin-Dna Adduct
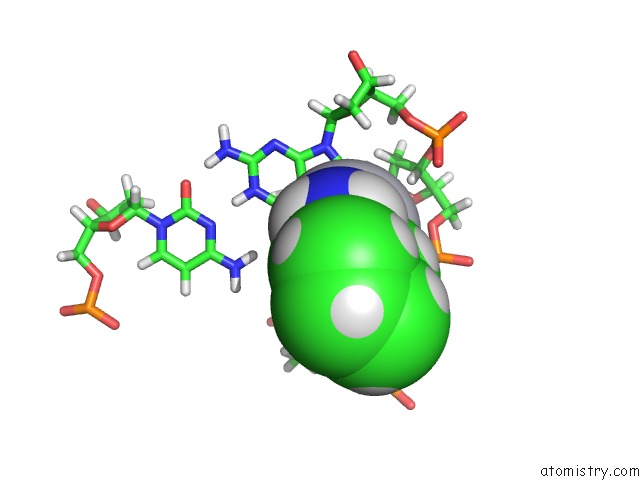
Mono view
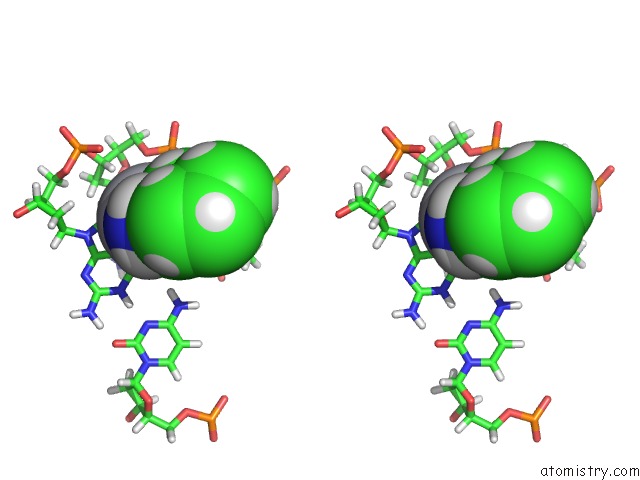
Stereo pair view

Mono view

Stereo pair view
A full contact list of Platinum with other atoms in the Pt binding
site number 1 of High Resolution Solution uc(Nmr) Structures of Oxaliplatin-Dna Adduct within 5.0Å range:
|
Reference:
D.Bhattacharyya,
S.Ramachandran,
S.Sharma,
W.Pathmasiri,
C.L.King,
I.Baskerville-Abraham,
G.Boysen,
J.A.Swenberg,
S.L.Campbell,
N.V.Dokholyan,
S.G.Chaney.
Flanking Bases Influence the Nature of Dna Distortion By Platinum 1,2-Intrastrand (Gg) Cross-Links. Plos One V. 6 23582 2011.
ISSN: ESSN 1932-6203
PubMed: 21853154
DOI: 10.1371/JOURNAL.PONE.0023582
Page generated: Thu Oct 10 10:43:27 2024
ISSN: ESSN 1932-6203
PubMed: 21853154
DOI: 10.1371/JOURNAL.PONE.0023582
Last articles
F in 4PYYF in 4PZH
F in 4PYX
F in 4PX5
F in 4PY4
F in 4PWL
F in 4PVU
F in 4PUW
F in 4PU9
F in 4PUU