Platinum »
PDB 5djm-5v6i »
5djm »
Platinum in PDB 5djm: Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione.
Enzymatic activity of Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione.
All present enzymatic activity of Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione.:
2.5.1.18;
2.5.1.18;
Protein crystallography data
The structure of Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione., PDB code: 5djm
was solved by
L.J.Parker,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 27.98 / 1.90 |
| Space group | C 1 2 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 77.517, 90.130, 68.943, 90.00, 97.85, 90.00 |
| R / Rfree (%) | 20.7 / 24 |
Platinum Binding Sites:
The binding sites of Platinum atom in the Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione.
(pdb code 5djm). This binding sites where shown within
5.0 Angstroms radius around Platinum atom.
In total 3 binding sites of Platinum where determined in the Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione., PDB code: 5djm:
Jump to Platinum binding site number: 1; 2; 3;
In total 3 binding sites of Platinum where determined in the Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione., PDB code: 5djm:
Jump to Platinum binding site number: 1; 2; 3;
Platinum binding site 1 out of 3 in 5djm
Go back to
Platinum binding site 1 out
of 3 in the Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione.
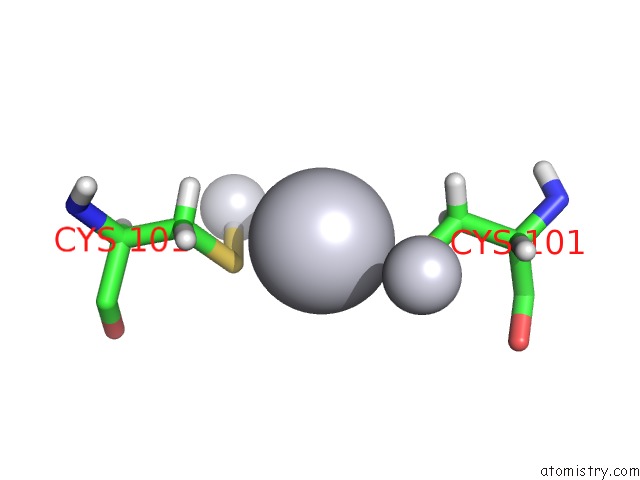
Mono view
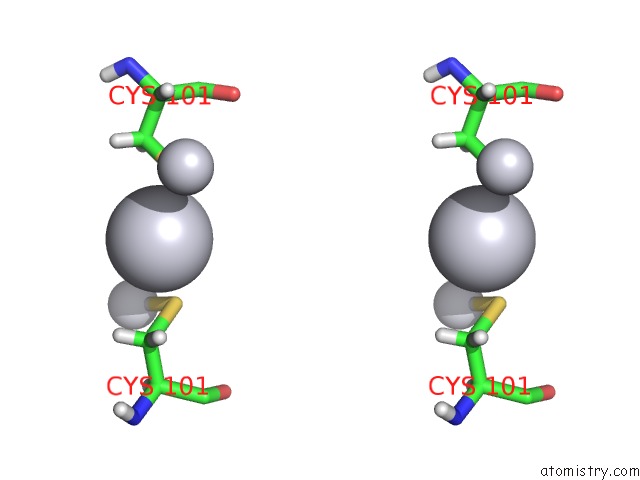
Stereo pair view

Mono view

Stereo pair view
A full contact list of Platinum with other atoms in the Pt binding
site number 1 of Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione. within 5.0Å range:
|
Platinum binding site 2 out of 3 in 5djm
Go back to
Platinum binding site 2 out
of 3 in the Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione.
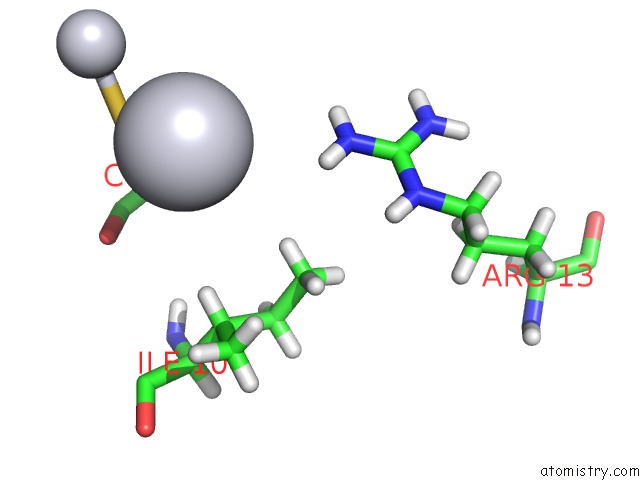
Mono view
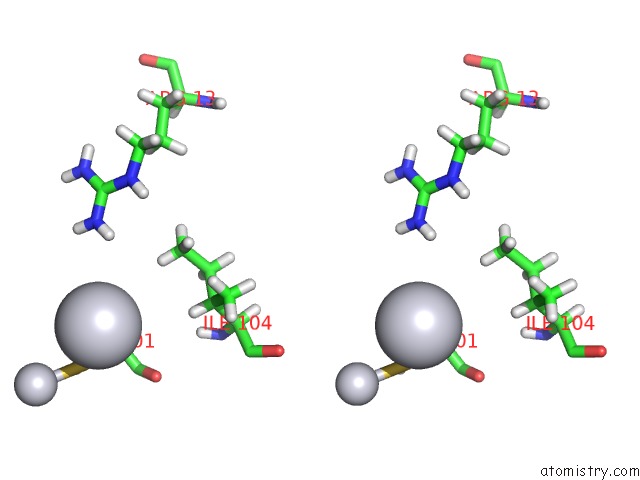
Stereo pair view

Mono view

Stereo pair view
A full contact list of Platinum with other atoms in the Pt binding
site number 2 of Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione. within 5.0Å range:
|
Platinum binding site 3 out of 3 in 5djm
Go back to
Platinum binding site 3 out
of 3 in the Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione.
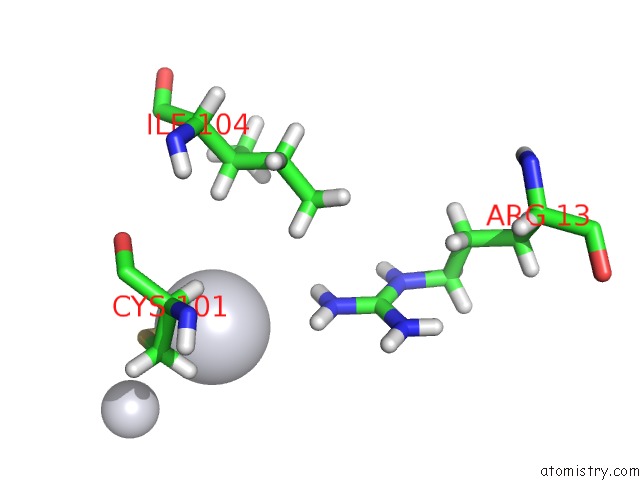
Mono view
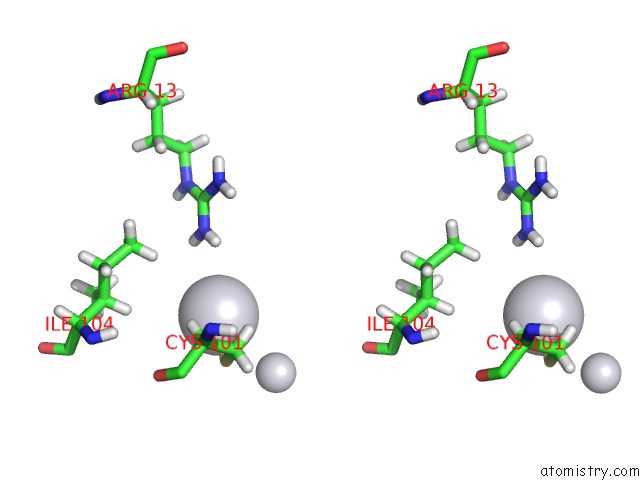
Stereo pair view

Mono view

Stereo pair view
A full contact list of Platinum with other atoms in the Pt binding
site number 3 of Structure of Wt Human Glutathione Transferase in Complex with Cisplatin in the Absence of Glutathione. within 5.0Å range:
|
Reference:
L.J.Parker,
L.C.Italiano,
N.C.Hancock,
J.Aitken,
H.H.Harris,
G.Hansen,
D.B.Ascher,
C.J.Morton,
M.W.Parker,
M.Lo Bello.
A Structure-Based Mechanism of Cisplatin Resistance Mediated By Glutathione Transferase P1-1 Proc.Natl.Acad.Sci.Usa 2019.
ISSN: ESSN 1091-6490
Page generated: Mon Aug 18 23:49:08 2025
ISSN: ESSN 1091-6490
Last articles
Sr in 3BNQSr in 2QJY
Sr in 2X53
Sr in 2SPT
Sr in 2XRM
Sr in 2WOH
Sr in 2RIO
Sr in 2PN4
Sr in 2QJK
Sr in 2QJP