Platinum »
PDB 1a2e-1sj7 »
1lzw »
Platinum in PDB 1lzw: Structural Basis of Clps-Mediated Switch in Clpa Substrate Recognition
Protein crystallography data
The structure of Structural Basis of Clps-Mediated Switch in Clpa Substrate Recognition, PDB code: 1lzw
was solved by
K.Zeth,
R.B.Ravelli,
K.Paal,
S.Cusack,
B.Bukau,
D.A.Dougan,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.69 / 2.50 |
| Space group | P 21 21 2 |
| Cell size a, b, c (Å), α, β, γ (°) | 90.880, 114.590, 38.490, 90.00, 90.00, 90.00 |
| R / Rfree (%) | 24.3 / 26.7 |
Platinum Binding Sites:
The binding sites of Platinum atom in the Structural Basis of Clps-Mediated Switch in Clpa Substrate Recognition
(pdb code 1lzw). This binding sites where shown within
5.0 Angstroms radius around Platinum atom.
In total only one binding site of Platinum was determined in the Structural Basis of Clps-Mediated Switch in Clpa Substrate Recognition, PDB code: 1lzw:
In total only one binding site of Platinum was determined in the Structural Basis of Clps-Mediated Switch in Clpa Substrate Recognition, PDB code: 1lzw:
Platinum binding site 1 out of 1 in 1lzw
Go back to
Platinum binding site 1 out
of 1 in the Structural Basis of Clps-Mediated Switch in Clpa Substrate Recognition

Mono view
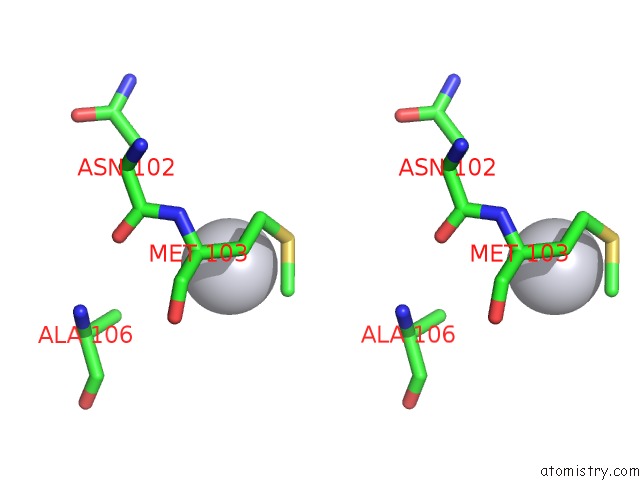
Stereo pair view

Mono view

Stereo pair view
A full contact list of Platinum with other atoms in the Pt binding
site number 1 of Structural Basis of Clps-Mediated Switch in Clpa Substrate Recognition within 5.0Å range:
|
Reference:
K.Zeth,
R.B.Ravelli,
K.Paal,
S.Cusack,
B.Bukau,
D.A.Dougan.
Structural Analysis of the Adaptor Protein Clps in Complex with the N-Terminal Domain of Clpa Nat.Struct.Biol. V. 9 906 2002.
ISSN: ISSN 1072-8368
PubMed: 12426582
DOI: 10.1038/NSB869
Page generated: Thu Oct 10 10:35:46 2024
ISSN: ISSN 1072-8368
PubMed: 12426582
DOI: 10.1038/NSB869
Last articles
Mg in 4KQSMg in 4KQ5
Mg in 4KQL
Mg in 4KPZ
Mg in 4KPX
Mg in 4KPF
Mg in 4KPE
Mg in 4KPJ
Mg in 4KOE
Mg in 4KPD